Content
Research overview

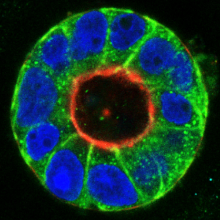
Tumor cells are characterized by complex molecular changes that lead to aberrant survival, growth and motility. Our team investigates the interplay of tumor suppressive and oncogenic signaling networks in cellular transformation. A particular focus is on the signaling pathways that contribute to cancer cell metastasis. Using 3D tissue culture models, we study the molecular interactions with the tumor cell environment, aiming to identify new therapeutic points of intervention. We further employ advanced genetic engineering approaches, primary (co)culture models in combination with single cell and state-of-the-art imaging techniques.
Selected Current Projects
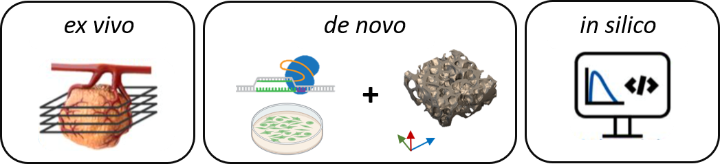
The 3R-US network develops a versatile tissue culture platform for drug testing to replace, reduce and refine (3R) animal experiments. Currently, most studies evaluating anti-cancer therapeutics proceed from human cell culture to preclinical cell line or patient derived mouse tumor models (CDX, PDX). These CDX/PDX models poorly recapitulate the complexity, heterogeneity, and physiology of human tumors. With interdisciplinary partners in cell biology, biomedical engineering and oncology, 3R-US will therefore advance methods that enable the long-term ex vivo cultivation of mouse and human tumor tissue slices. New (targeted) therapeutics and combination therapies will be evaluated in these slice cultures and the mechanisms of drug action investigated. Furthermore, 3R-US will develop de novo human tumor models using 3D printing to study heterogeneous cell interactions and drug responses. Finally, 3R-US will use the data derived from the ex vivo and de novo approaches to establish and validate in silico tumor models that reliably predict the pharmacokinetics and therapy response to anti-cancer drugs.
https://www.verbund.uni-stuttgart.de/3r-biomedicus/3r-research/3r-us/
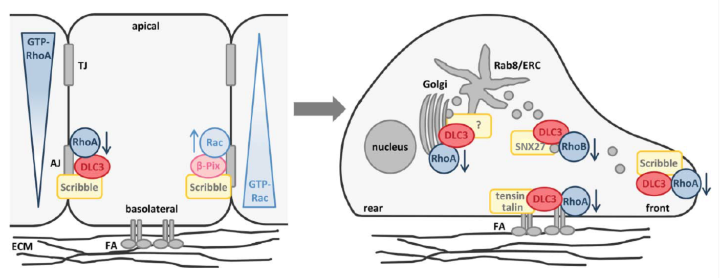
The Rho GTPase activating (GAP) protein Deleted in Liver Cancer 1 (DLC1) has emerged as an important tumor suppressor, whose downregulation in various types of cancer may be as common as that of p53. Our work has shown that DLC1 loss facilitates the aberrant migration of breast cancer cells, whereas expression of the family member DLC3 is crucial for the establishment and maintenance of cell polarity and cell-cell adhesions. While Rho signaling is regulated negatively by GAP proteins on the one hand, it is regulated positively by GEF proteins on the other. We are especially interested in unraveling how specific GEF-GAP networks regulate spatiotemporal Rho signaling and how dysregulation contributes to the metastatic behavior of cancer cells. As partner within the H2020 SECRET Innovative Training Network, we aim to identify drug vulnerabilities of cancer cells with DLC downregulation.
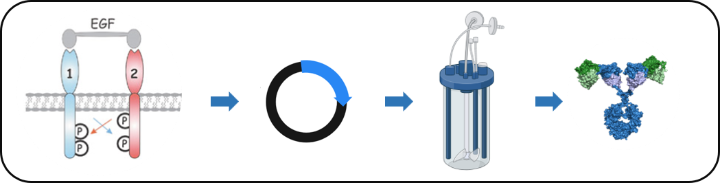
The ErbB family of receptor tyrosine kinases (RTKs) plays a well-established role in cancer development and progression. ErbB2/HER2, for example, is amplified and/or overexpressed in approximately 25% of breast cancer patients, correlating with poor clinical prognosis, whereas EGFR/ErbB1 expression is elevated in 70% of triple-negative breast cancers. Although these receptors are prime targets for pharmacological intervention in the clinic, cancer cells are often non-responsive or become resistant to treatment. In collaboration with the lab of Prof. Roland Kontermann we are developing novel RTK-based targeted approaches for personalized treatment of solid cancers.
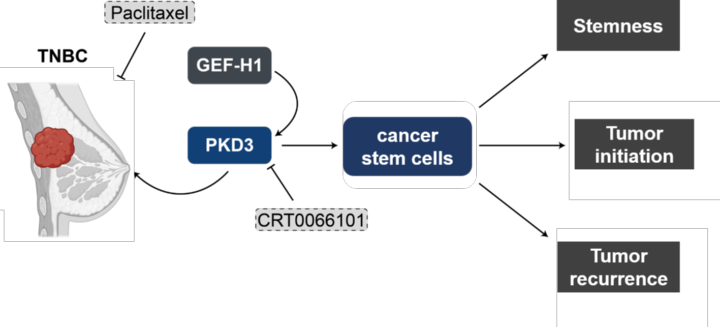
Triple-negative breast cancer (TNBC) is an especially aggressive form of breast cancer for which targeted therapies are still in their infancy. We identified the serine-threonine kinase PKD3 to be upregulated in TNBC where it drives proliferation, cell motility and cancer stem cell survival. By mapping the PKD3 interactome and kinome our goal is to identify druggable signaling nodes for this cancer type.
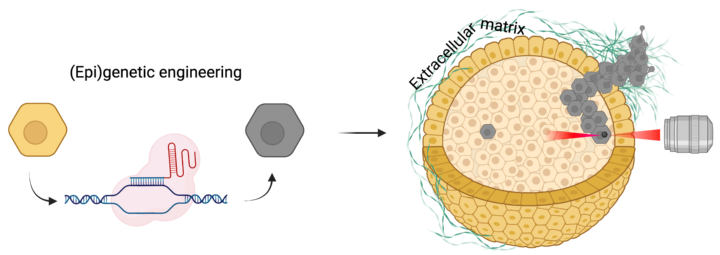
Loss of tumor suppressors and activation of oncogenes are key events in cancer development. While these events can spontaneously occur in multiple cells, only a subset of these cells will eventually contribute to tumor formation or promote metastasis. The conditions that facilitate this, particularly in 3D environments in which cells typically grow within tissues, are poorly understood. This interdisciplinary project tackles this topic by combining state of the art CRISPR/Cas9 epigenetic editing, cancer cell biology and biophysical methodologies. This project is performed in close collaboration with the Guo lab of cell mechanics (MIT) and is funded by the MISTI Global Seed Fund – University of Stuttgart program.
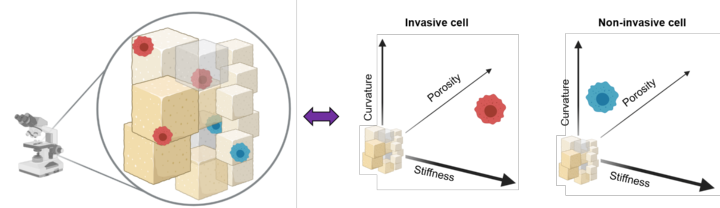
More than 70% of breast cancer associated deaths hail from the metastatic spread of the disease to distant organs. Interestingly, most breast cancer metastases affect the bones, the mechanisms involved in this tissue-specific enrichment being still poorly understood. In particular, the role of biophysical cues such as tissue topography and stiffness in this process are not clear. In this interdisciplinary project we will develop high resolution bone mimetic scaffolds to assay the metastatic colonization process. This work is performed together with the Heymann and Hörning groups from the IBBS. The initiative is funded by the Research Seed Capital Program (MWK Baden-Württemberg & University of Stuttgart).
Our Partners
We work in close collaboration with Prof. Matthias Schwab (Margarete Fischer Bosch Institute of Clinical Pharmacology, Stuttgart), Prof. Walter Aulitzky (Robert Bosch Hospital, Stuttgart), Prof. Christine Sers (Charité, Berlin), Prof. Tilman Brummer and Prof. Melanie Boerries (University of Freiburg), Prof. Hauke Busch (University of Luebeck), and Prof. Boris Macek (Proteome Center Tübingen).
Publications
Click here for a full publication list.
News
The prize was awarded for her outstanding contribution to the manuscript: Repeat DNA methylation is modulated by adherens junction signaling., published in Communications Biology (Nature Portfolio).
The work is a collaborative achievement by researchers from the Olayioye group (IZI), Jeltsch group (Institute of Biochemistry, IBC), and the Massachusetts Institute of Technology (Department of Mechanical Engineering).
Since 2018, the University of Stuttgart has honored excellent scientific publications from each of its ten faculties as part of its annual Day of Research event. Each award is endowed with €2,500.
On July 9 starting at 4 PM, the awardees from the faculties will present their publications in short, entertaining talks.
Where? Campus Vaihingen, Room 38.02, Universitätsstraße 38, 70569 Stuttgart.
Optional: Want to know more about the interface between Cell Signaling and Epigenetics, check out our recently funded EpiSignal graduate college (GRK).
Integrated mathematical and experimental modeling uncovers enhanced EMT plasticity upon loss of the DLC1 tumor suppressor
Sebastian Höpfl, Merih Özverin, Helena Nowack, Raluca Tamas, Andrew G. Clark, Nicole Radde, Monilola A. Olayioye
Abstract
Epithelial-mesenchymal transition (EMT) plays an essential role in embryonic development, wound healing, and tumor progression. Partial EMT states have been linked to metastatic dissemination and drug resistance. Several interconnected feedback loops at the RNA and protein levels control the transition between different cellular states. Using a combination of mathematical modeling and experimental analyses in the TGFβ-responsive breast epithelial MCF10A cell model, we identify a central role for the tumor suppressor protein Deleted in Liver Cancer 1 (DLC1) during EMT. By extending a previous model of EMT comprising key transcription factors and microRNAs, our work shows that DLC1 acts as a positive regulator of TGFβ-driven EMT, mainly by promoting SNAIL1 expression. Our model predictions indicate that DLC1 loss impairs EMT progression. Experimental analyses confirm this prediction and reveal the acquisition of a partial EMT phenotype in DLC1-depleted cells. Furthermore, our model results indicate a possible EMT reversion to partial or epithelial states upon DLC1 loss in MCF10A cells induced toward mesenchymal phenotypes. The increased EMT plasticity of cells lacking DLC1 may explain its importance as a tumor suppressor.
Reciprocal regulation of Solo and Src orchestrates Src trafficking to promote mesenchymal cell migration
Florian Meyer, Cristiana Lungu, Bettina Noll, David Benz, Felix Fränkle, Miguel Â. Ferreira, Raluca Tamas, Monilola A. Olayioye
Highlights:
- Solo expression is increased in breast cancer cells with high EMT signatures.
- Solo promotes breast cancer cell migration in a RhoGEF-dependent manner.
- Src phosphorylates Solo at Y242 and cooperates with Solo to promote cell migration.
- Loss of Solo disrupts Src trafficking and focal adhesion signaling dynamics.
Summary
Rho GTPases are key regulators of cell motility and membrane trafficking, influencing critical processes such as epithelial-mesenchymal transition (EMT). Among them, the small GTPase RhoB plays a pivotal role, but the mechanisms underlying its regulation remain largely unclear. We have previously identified the Rho guanine nucleotide exchange factor (RhoGEF) Solo (ARHGEF40) as a regulator of endosomal RhoB in epithelial cells. Here, we find that Solo is upregulated in breast cancer cells with high EMT scores and promotes cell motility through its RhoGEF activity. Solo’s ability to enhance migration is further regulated by phosphorylation at tyrosine 242, mediated by the proto-oncogene Src. By combining high-resolution imaging with photoconversion assays, we further demonstrate that Solo regulates Src trafficking dynamics, localization, and consequently signaling at focal adhesions. Together, our data identify Solo as a novel feedback regulator of Src and a key driver of the motility of breast cancer cells with mesenchymal characteristics.
Congratulations to Lisa Brenner on the successful defense of her PhD thesis! Using a powerful combination of CRISPR/Cas9, high-resolution imaging, and cell biology, Lisa uncovered a novel pathway that bridges epigenetics and cell signaling. Her research revealed that in confluent breast epithelial cell monolayers, DNA methylation of α-satellite repeats is reduced compared to low-density cultures—a change linked with increased transcriptional activity. She also demonstrated that the junctional protein E-cadherin is essential for transmitting signals via the actin cytoskeleton into the nucleus. Notably, this pathway is disrupted in cancer cells lacking E-cadherin, opening new avenues to explore how the loss of contact inhibition contributes to tumor development.
For more details, check out our recent publication in Communications Biology: Repeat DNA methylation is modulated by adherens junction signaling | Communications Biology
and the accompanying “Behind the Paper” blog post on Springer Nature’s Research Communities: Research Communities by Springer Nature
We applaud Lisa’s achievements and wish her every success as she embarks on her new role as Scientific Project Manager at the Center for Personalized Medicine, University Hospital Tübingen.
Does that sound like an exciting research topic? We are looking for talent to join our teams within the newly funded EpiSignal graduate school: https://www.grk3112.uni-stuttgart.de/
Dr. Cristiana Lungu has been invited to give a presentation at Cell Bio 2024, the joint meeting of the ASCB and EMBO, taking place in San Diego from December 14–18, 2024. The talk will showcase the use of opts-proteomics, an advanced spatial proteomics technology, to obtain insights into the misregulation of epigenetic proteins in cancer cells.
Cell Bio 2024 is a premier event in the field of cell biology, bringing together leading researchers to explore foundational and interdisciplinary topics: https://www.ascb.org/cellbio2024/program/
Authors: Yannick Frey, Cristiana Lungu, Monilola A. Olayioye
Cell Signal. 2024 Nov 14:111505, DOI: 10.1016/j.cellsig.2024.111505
Abstract
The DLC (Deleted in Liver Cancer) family of RhoGAP (Rho GTPase-activating) proteins has been extensively studied since the identification of the first family member nearly 30 years ago. Rho GTPase signaling is essential for various cellular processes, including cytoskeletal dynamics, cell migration, and proliferation. Members of the DLC family are key regulators of this signaling pathway, with well-established roles in development and carcinogenesis. Here, we provide a comprehensive review of research into DLC regulation and cellular functions over the last three decades. In particular, we summarize control mechanisms of DLC gene expression at both the transcriptional and post-transcriptional level. Additionally, recent advances in understanding the post-translational regulation of DLC proteins that allow for tuning of protein activity and localization are highlighted. This detailed overview will serve as resource for future studies aimed at further elucidating the complex regulatory mechanisms of DLC family proteins and exploring their potential as targets for therapeutic applications.
Monilola Olayioye (Lab Head)
Cristiana Lungu (Postdoctoral Scientist and Project Leader)
Raluca Tamas (3R-BioMedicUS Scientific Coordinator)
Bettina Noll (Postdoctoral Scientist)
Camille Dantzer (Postdoctoral Scientist)
Merih Özverin (PhD student)
Florian Meyer (PhD student)
Stella Asmanidou (joint PhD student with the Kontermann lab)
Fiona Kühnel (PhD student)
Ann-Kathrin Löffler (joint PhD student with the Kontermann lab)
Dimitra Chrysouli (Bachelor student)
Manuel Baumann (Master student)
Saskia Ebert (Master student)
Tim Bucher (Bachelor student)
Nina Pelzer (student assistant)
Ronja Schiffler (student assistant)
Simone Schmid (Technician)
Ilaria Martorana (Secretary)
How to join our Group
Undergraduate students interested in conducting a BSc/MSc research project should get in touch at an early stage with their CV as space is limited. PhD and postdoctoral positions are advertised on our career portal JoinUS. We are happy to support highly qualified and competitive candidates that wish to apply for fellowship funding programs to join our research team.